
Interpreting a leaf identification model with LIME
This notebook demonstrates the use of DIANNA with the LIME method on the leafsnap30 image dataset. A pre-trained neural network classifier is used, which identifies the species of leaf based on an image of it.
LIME (Local Interpretable Model-agnostic Explanations) is an explainable-AI method that aims to create an interpretable model that locally represents the classifier. For more details see the LIME paper.
NOTE: This tutorial is still work-in-progress, the final results need to be improved by tweaking the LIME parameters
Colab Setup
[1]:
running_in_colab = 'google.colab' in str(get_ipython())
if running_in_colab:
# install dianna
!python3 -m pip install dianna[notebooks]
# download data used in this demo
import os
base_url = 'https://raw.githubusercontent.com/dianna-ai/dianna/main/dianna/'
paths_to_download = ['./data/leafsnap_example_acer_rubrum.jpg', './labels/leafsnap_classes.csv', './models/leafsnap_model.onnx']
for path in paths_to_download:
!wget {base_url + path} -P {os.path.dirname(path)}
0 - Imports and paths
[1]:
from PIL import Image
import matplotlib.pyplot as plt
import numpy as np
from pathlib import Path
import dianna
from dianna import visualization
[2]:
true_species = 'acer_rubrum'
image_path = Path('..','..','..','dianna','data', f'leafsnap_example_{true_species}.jpg')
model_path = Path('..','..','..','dianna','models', 'leafsnap_model.onnx')
model_classes_path = Path('..','..','..','dianna','labels' ,'leafsnap_classes.csv')
1 - Loading the data
Two files are loaded here: 1. A file containg the numerical index that belongs to each class (=species of leaf). This is used so we know which output of the neural network corresponds to the class we want to run LIME on. 2. An image of a leaf, belonging to the Acer Rubrum species.
DIANNA requires input in numpy format, so the .jpg image is loaded and converted into a numpy array. The pixel values are then converted to the 0-1 range, which the classifier requires. Finally, DIANNA requires the presence of a batch axis, and it needs to know where the batch and colour channel axes are located in the input data.
[3]:
# load the model class definitions
class_to_idx = dict(np.genfromtxt(model_classes_path, dtype=None, encoding=None, delimiter=','))
true_species = 'acer_rubrum'
true_species_index = class_to_idx[true_species]
[4]:
# load and plot the example image
img = np.array(Image.open(Path('..','..','..','dianna','data', f'leafsnap_example_{true_species}.jpg')))
plt.imshow(img)
plt.title(f'Species: {true_species}');
# the model expects float32 values in the 0-1 range for each pixel, with the colour channels as first axis
# the .jpg file has 0-255 ints with the channel axis last so it needs to be changed
input_data = img.transpose(2, 0, 1).astype(np.float32) / 255.
# define axis labels. Required is the channels axis
# in this example image, the channels axis is the first axis
axis_labels = {0: 'channels'}
2 - Applying LIME with DIANNA
The simplest way to run DIANNA on image data is with dianna.explain_image. The arguments are: * The path to the model in ONNX format * The image we want to explain * The name of the explainable-AI method we want to use, here LIME * The location of the batch and colour channel axes in the data * The numerical indices of the classes we want an explanation for
[5]:
# An explanation is returned for each label, but we ask for just one label so the output is a list of length one.
explanation_heatmap = dianna.explain_image(model_path, input_data, 'LIME', axis_labels=axis_labels, labels=[true_species_index])[0]
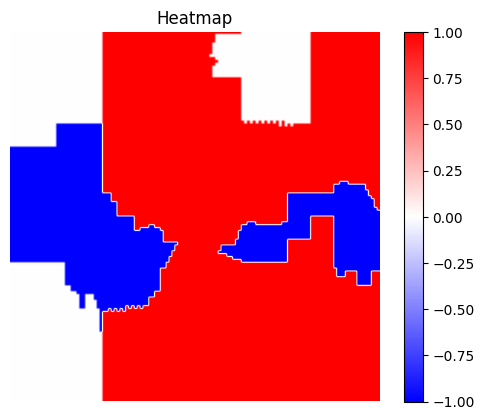
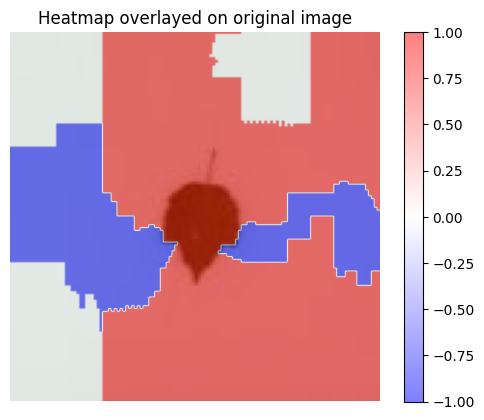
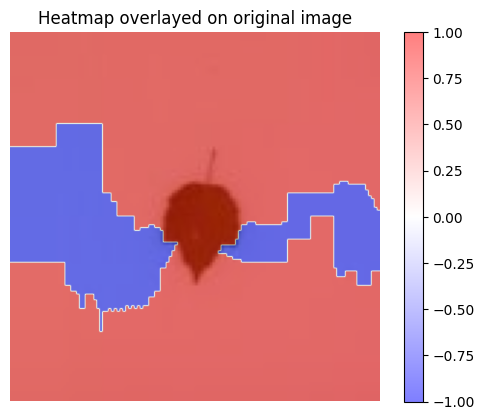
3 - Visualization
[19]:
visualization.plot_image(explanation_heatmap, heatmap_cmap='bwr')
plt.title('Heatmap')
plt.axis('off')
visualization.plot_image(explanation_heatmap, original_data=img, heatmap_cmap='bwr')
plt.title('Heatmap overlayed on original image')
plt.axis('off')
plt.show()
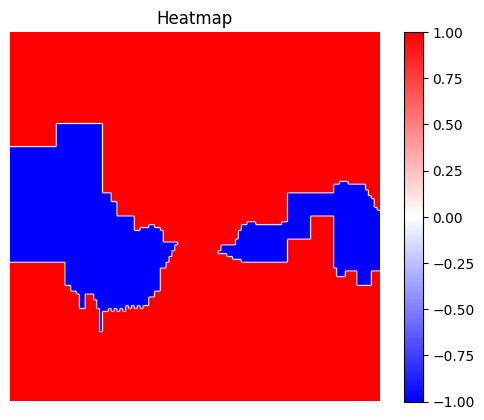
dianna.explain_image and will automatically be used by LIME.[16]:
explanation_heatmap_customized = dianna.explain_image(model_path, input_data, 'LIME', axis_labels=axis_labels, labels=[true_species_index],
num_features=30, num_samples=1000)[0]
[17]:
visualization.plot_image(explanation_heatmap_customized, heatmap_cmap='bwr')
plt.title('Heatmap')
plt.axis('off')
visualization.plot_image(explanation_heatmap_customized, original_data=img, heatmap_cmap='bwr')
plt.title('Heatmap overlayed on original image')
plt.axis('off')
plt.show()
[ ]: